Advanced: Sparse Matrices for Large-Scale Networks
Author
Sadamori Kojaku
Published
August 14, 2025
1 The Scale Problem: From Königsberg to Global Networks
Modern networks have billions of nodes—far beyond Königsberg’s 4 landmasses. A dense adjacency matrix for Earth’s 8 billion people would require 512 exabytes of memory!
Real networks are sparse: most node pairs aren’t connected. Edge lists save memory but are inefficient for common operations like finding neighbors or computing degrees.
Think about the following operations:
Degree: How many friends does a person have?
Neighbors: Who are the friends of a person?
These operations are very common in network analysis. To do so, you need to go through all the edges in the network. This is not efficient, especially for large networks.
2 Solution: Sparse Matrices
We say a matrix is sparse if the matrix has only a handful of non-zero entries. This is indeed the case for most real-world networks. For such networks, we can use a special type of data type called Compressed Sparse Row (CSR) or Compressed Sparse Column (CSC) to represent the network. This is widely used in many network analysis tools and makes it possible to process large networks in practice.
Here is a short video explaining the CSR format. Good one to watch if you are visual learner.
Figure 1: A sparse matrix is a matrix with only a handful of non-zero entries represented with a compressed sparse frmat.
CSR stores only non-zero matrix entries using three arrays, making it the standard for large-scale network analysis.
Understanding CSR Structure
CSR format uses three arrays to represent a sparse matrix:
data: Contains all non-zero values
indices: Column indices of each non-zero value
indptr: Row pointers indicating where each row starts in the data array
Memory efficiency: For a sparse matrix with m non-zero entries out of n^2 total entries, CSR uses O(m + n) memory instead of O(n^2). For social networks where m \ll n^2, this is a massive saving!
import numpy as npfrom scipy import sparse# Create a small example network# Let's represent the same 5-node network from earlierdense_matrix = np.array([ [0, 1, 1, 0, 0], # Node 0 connects to nodes 1, 2 [1, 0, 1, 1, 0], # Node 1 connects to nodes 0, 2, 3 [1, 1, 0, 0, 1], # Node 2 connects to nodes 0, 1, 4 [0, 1, 0, 0, 1], # Node 3 connects to nodes 1, 4 [0, 0, 1, 1, 0] # Node 4 connects to nodes 2, 3])# Convert to CSR formatcsr_matrix = sparse.csr_matrix(dense_matrix)print("Dense matrix shape:", dense_matrix.shape)print("CSR matrix shape:", csr_matrix.shape)print("Non-zero entries:", csr_matrix.nnz)print("Memory saved: {:.1f}%".format((1- csr_matrix.nnz / dense_matrix.size) *100))
CSR format integrates seamlessly with our previously defined functions:
def is_walk_sparse(sequence, csr_matrix):""" Check if a sequence forms a valid walk using sparse CSR matrix. """iflen(sequence) <2:returnTrue sequence = np.array(sequence) current_nodes = sequence[:-1] next_nodes = sequence[1:]# Use CSR matrix indexing - still works with advanced indexing! edges_exist = csr_matrix[(current_nodes, next_nodes)]# Convert sparse result to array and checkreturn np.all(edges_exist ==1)# Test with our CSR matrixtest_walk = [0, 1, 2, 4, 3, 1]print(f"Walk {test_walk} is valid: {is_walk_sparse(test_walk, csr_matrix)}")
Walk [0, 1, 2, 4, 3, 1] is valid: True
Best Practices
When to use CSR: - Sparse matrices - Row-based operations (computing degrees, finding neighbors) - Matrix-vector multiplication
When to use dense matrices: - Small networks - Dense networks - Frequent random access to individual entries
There are several different sparse matrix formats such as COO, CSC, and LIL. If you are interested in learning more, you can check out this blog post by Matt Eding. The following video is also a good one to watch.
Advanced CSR Features
# Submatrix extraction - get connections for subset of nodessubset_nodes = [0, 1, 2]subgraph = csr_matrix[subset_nodes][:, subset_nodes]print("Subgraph for nodes [0, 1, 2]:")print(subgraph.toarray())# Efficient boolean operations# Find nodes with degree > 2high_degree_nodes = np.where(degrees >2)[0]print("High degree nodes (> 2 connections):", high_degree_nodes)# Matrix powers for path countingpaths_3 = csr_matrix **3# Counts 3-step pathsprint("3-step path counts:")print(paths_3.toarray())
The CSR format transforms network analysis from impossible to practical for large-scale networks. By storing only the essential information (non-zero connections), we can analyze networks with millions of nodes and billions of edges on standard hardware—something that would require exabytes of memory with dense matrices!
---title: "Advanced: Sparse Matrices for Large-Scale Networks"jupyter: python3execute: enabled: true---## The Scale Problem: From Königsberg to Global NetworksModern networks have billions of nodes—far beyond Königsberg's 4 landmasses. A dense adjacency matrix for Earth's 8 billion people would require 512 exabytes of memory!Real networks are **sparse**: most node pairs aren't connected. Edge lists save memory but are inefficient for common operations like finding neighbors or computing degrees.::: {.column-margin}Think about the following operations:- **Degree**: How many friends does a person have?- **Neighbors**: Who are the friends of a person?These operations are very common in network analysis. To do so, you need to go through **all the edges** in the network. This is not efficient, especially for large networks.:::## Solution: Sparse MatricesWe say a matrix is sparse if the matrix has only a handful of non-zero entries. This is indeed the case for most real-world networks. For such networks, we can use a special type of data type called **Compressed Sparse Row (CSR)** or **Compressed Sparse Column (CSC)** to represent the network. This is widely used in many network analysis tools and makes it possible to process large networks in practice.To learn more, [here is a very good blog post by Matt Eding about efficient network representations](https://matteding.github.io/2019/04/25/sparse-matrices/).## SciPy's Compressed Sparse Row (CSR) Format::: {.column-margin}Here is a short video explaining the CSR format. Good one to watch if you are visual learner.<iframe width="250" height="150" src="https://www.youtube.com/embed/a2LXVFmGH_Q?si=F25ylU_vMHkQ0SP1" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture; web-share" referrerpolicy="strict-origin-when-cross-origin" allowfullscreen></iframe>:::::: {#fig-csr}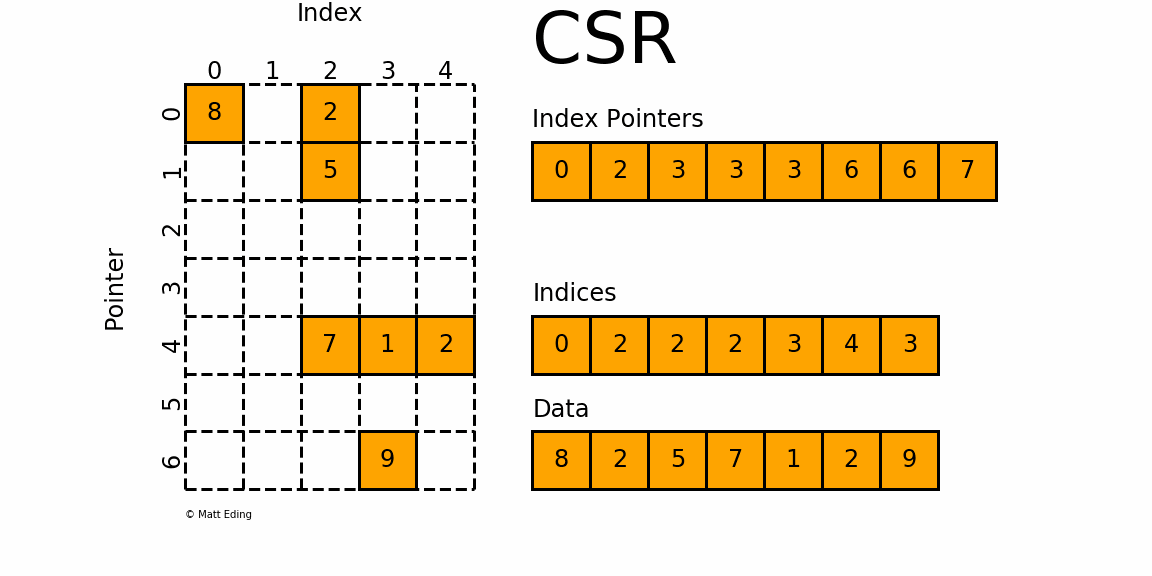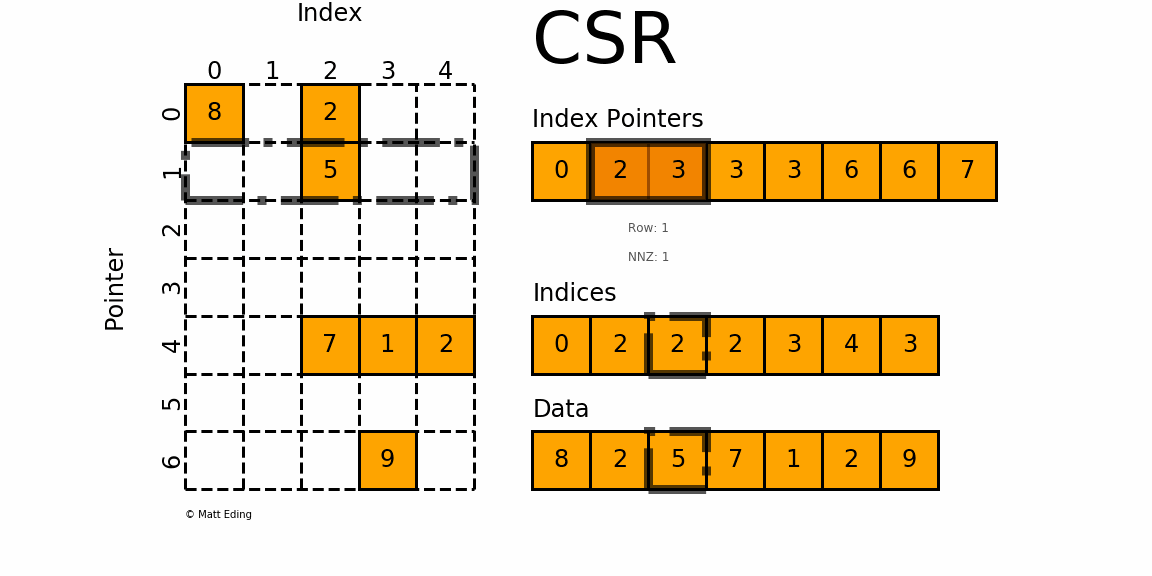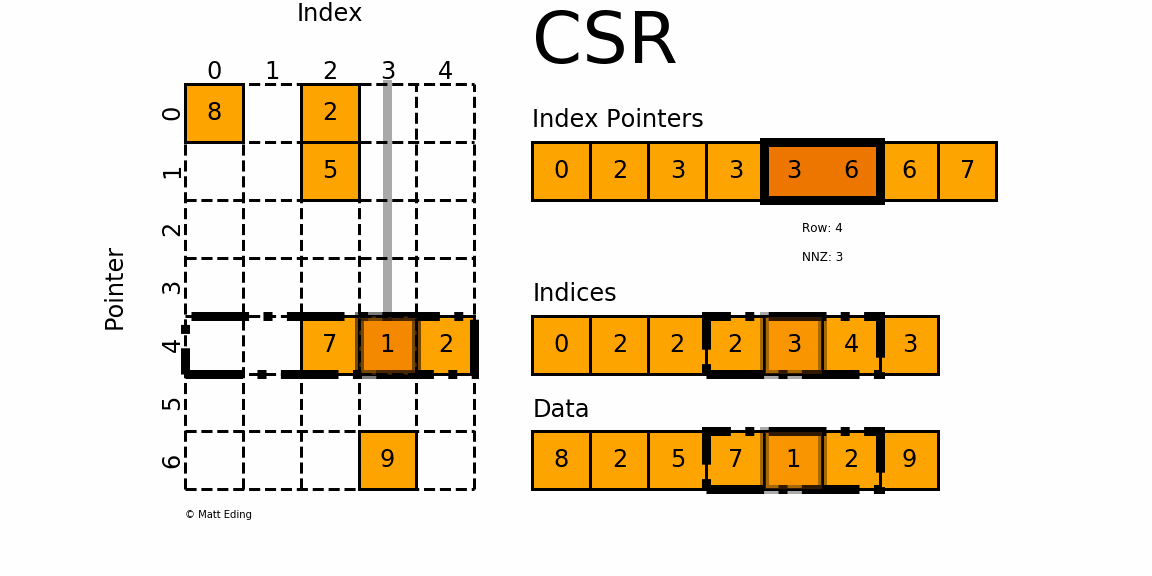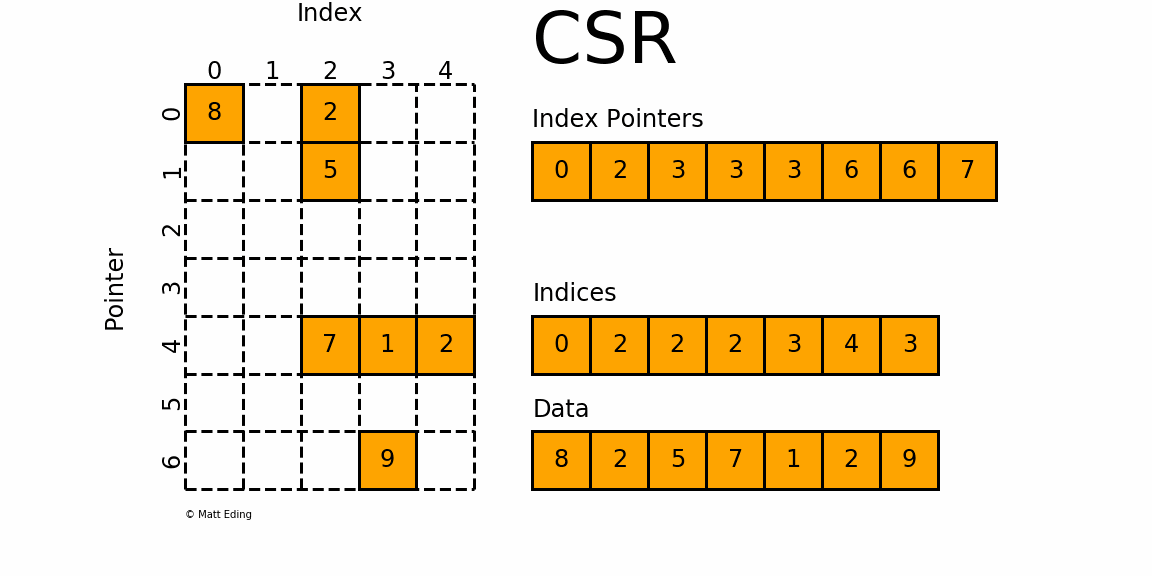A sparse matrix is a matrix with only a handful of non-zero entries represented with a compressed sparse frmat.:::CSR stores only non-zero matrix entries using three arrays, making it the standard for large-scale network analysis.### Understanding CSR StructureCSR format uses three arrays to represent a sparse matrix:1. **`data`**: Contains all non-zero values2. **`indices`**: Column indices of each non-zero value3. **`indptr`**: Row pointers indicating where each row starts in the data array::: {.column-margin}**Memory efficiency**: For a sparse matrix with $m$ non-zero entries out of $n^2$ total entries, CSR uses $O(m + n)$ memory instead of $O(n^2)$. For social networks where $m \ll n^2$, this is a massive saving!:::```{python}import numpy as npfrom scipy import sparse# Create a small example network# Let's represent the same 5-node network from earlierdense_matrix = np.array([ [0, 1, 1, 0, 0], # Node 0 connects to nodes 1, 2 [1, 0, 1, 1, 0], # Node 1 connects to nodes 0, 2, 3 [1, 1, 0, 0, 1], # Node 2 connects to nodes 0, 1, 4 [0, 1, 0, 0, 1], # Node 3 connects to nodes 1, 4 [0, 0, 1, 1, 0] # Node 4 connects to nodes 2, 3])# Convert to CSR formatcsr_matrix = sparse.csr_matrix(dense_matrix)print("Dense matrix shape:", dense_matrix.shape)print("CSR matrix shape:", csr_matrix.shape)print("Non-zero entries:", csr_matrix.nnz)print("Memory saved: {:.1f}%".format((1- csr_matrix.nnz / dense_matrix.size) *100))```### Inside the CSR FormatLet's examine the internal structure of our CSR matrix:```{python}print("CSR internal arrays:")print("data (non-zero values):", csr_matrix.data)print("indices (column positions):", csr_matrix.indices)print("indptr (row pointers):", csr_matrix.indptr)# Let's trace through how CSR worksprint("\nDecoding CSR structure:")for i inrange(len(csr_matrix.indptr) -1): start = csr_matrix.indptr[i] end = csr_matrix.indptr[i +1] row_data = csr_matrix.data[start:end] row_indices = csr_matrix.indices[start:end]print(f"Row {i}: values {row_data} at columns {row_indices}")```::: {.callout-note title="How CSR Works"}For row $i$, the non-zero values are stored in `data[indptr[i]:indptr[i+1]]` with their column positions in `indices[indptr[i]:indptr[i+1]]`.For example, if `indptr[0] = 0` and `indptr[1] = 2`, then row 0 has non-zero values `data[0:2]` at columns `indices[0:2]`.:::### Creating CSR Matrices from Edge ListsThe most common way to create network CSR matrices is from edge lists:```{python}# Define our network as an edge listedges = [ (0, 1), (0, 2), # Node 0 connections (1, 2), (1, 3), # Node 1 connections (2, 4), # Node 2 connections (3, 4) # Node 3 connections]# Extract source and target nodessources = [edge[0] for edge in edges]targets = [edge[1] for edge in edges]# For undirected graphs, add reverse edgesall_sources = sources + targetsall_targets = targets + sources# Create data array (all ones for unweighted graph)data_values = np.ones(len(all_sources))# Create CSR matrix directly from edge listn_nodes =5csr_from_edges = sparse.csr_matrix( (data_values, (all_sources, all_targets)), shape=(n_nodes, n_nodes))print("CSR from edge list:")print(csr_from_edges.toarray())```### Efficient Operations with CSRCSR format enables efficient network operations that would be slow with dense matrices:```{python}# Node degrees - sum each rowdegrees = np.array(csr_matrix.sum(axis=1)).flatten()print("Node degrees:", degrees)# Find neighbors of node 1node_1_neighbors = csr_matrix.indices[csr_matrix.indptr[1]:csr_matrix.indptr[2]]print("Node 1 neighbors:", node_1_neighbors)# Matrix multiplication for 2-hop pathstwo_hop_matrix = csr_matrix @ csr_matrixprint("Two-hop connections (shows paths of length 2):")print(two_hop_matrix.toarray())```::: {.column-margin}**Matrix multiplication**: `@` is the matrix multiplication operator in SciPy and NumPy. It is equivalent to `np.dot`.:::### Memory Comparison: Dense vs CSRLet's demonstrate the memory efficiency with a larger, sparser network:```{python}# Create a larger sparse networkn =1000density =0.01# Only 1% of edges exist# Generate random sparse matrixnp.random.seed(42)large_dense = sparse.random(n, n, density=density, format='csr')print(f"Network size: {n} × {n} = {n**2:,} potential edges")print(f"Actual edges: {large_dense.nnz:,}")print(f"Sparsity: {(1- large_dense.nnz / (n*n)) *100:.1f}% zeros")print(f"CSR memory usage: ~{(large_dense.nnz *2+ n) *4/1024:.1f} KB")print(f"Dense memory usage: ~{n*n *4/1024:.1f} KB")print(f"Memory savings: {((n*n *4) - (large_dense.nnz *2+ n) *4) / (n*n *4) *100:.1f}%")```### CSR for Network Analysis AlgorithmsCSR format integrates seamlessly with our previously defined functions:```{python}def is_walk_sparse(sequence, csr_matrix):""" Check if a sequence forms a valid walk using sparse CSR matrix. """iflen(sequence) <2:returnTrue sequence = np.array(sequence) current_nodes = sequence[:-1] next_nodes = sequence[1:]# Use CSR matrix indexing - still works with advanced indexing! edges_exist = csr_matrix[(current_nodes, next_nodes)]# Convert sparse result to array and checkreturn np.all(edges_exist ==1)# Test with our CSR matrixtest_walk = [0, 1, 2, 4, 3, 1]print(f"Walk {test_walk} is valid: {is_walk_sparse(test_walk, csr_matrix)}")```::: {.callout-tip title="Best Practices"}**When to use CSR**:- Sparse matrices- Row-based operations (computing degrees, finding neighbors)- Matrix-vector multiplication**When to use dense matrices**:- Small networks- Dense networks- Frequent random access to individual entries:::::: {.column-margin}There are several different sparse matrix formats such as COO, CSC, and LIL. If you are interested in learning more, you can check out [this blog post by Matt Eding](https://matteding.github.io/2019/04/25/sparse-matrices/). The following video is also a good one to watch.<iframe width="250" height="150" src="https://www.youtube.com/embed/SRgFScHYTy4?si=C75vxEnUjuLCerWF" title="YouTube video player" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture; web-share" referrerpolicy="strict-origin-when-cross-origin" allowfullscreen></iframe>:::### Advanced CSR Features```{python}# Submatrix extraction - get connections for subset of nodessubset_nodes = [0, 1, 2]subgraph = csr_matrix[subset_nodes][:, subset_nodes]print("Subgraph for nodes [0, 1, 2]:")print(subgraph.toarray())# Efficient boolean operations# Find nodes with degree > 2high_degree_nodes = np.where(degrees >2)[0]print("High degree nodes (> 2 connections):", high_degree_nodes)# Matrix powers for path countingpaths_3 = csr_matrix **3# Counts 3-step pathsprint("3-step path counts:")print(paths_3.toarray())```The CSR format transforms network analysis from impossible to practical for large-scale networks. By storing only the essential information (non-zero connections), we can analyze networks with millions of nodes and billions of edges on standard hardware—something that would require exabytes of memory with dense matrices!